KG-COVID-19
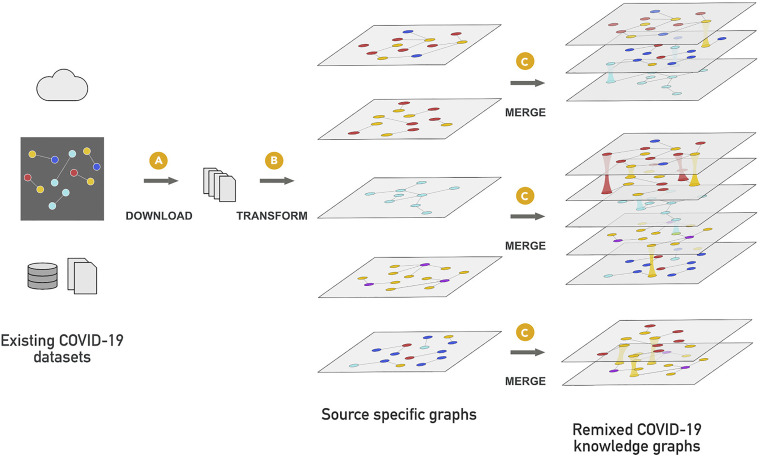
Introduction
KG-COVID-19 is a flexible framework that ingests and integrates heterogeneous biomedical data to produce knowledge graphs (KGs), and applied it to create a KG for COVID-19 response. This KG framework also can be applied to other problems in which siloed biomedical data must be quickly integrated for different research applications, including future pandemics.
Relation types
The challenge we suggest is prediction of drug targets, which are represented in the COVID-19 knowledge graph as:
drug (biolink:Drug) -- biolink:molecularly_interacts_with || biolink:interacts_with → target (biolink:Protein | biolink:Gene)of which there are currently 3372 (see query below)
Data sources and contact
- Github: https://github.com/Knowledge-Graph-Hub/kg-covid-19
- Contact and discussion: https://github.com/Knowledge-Graph-Hub/kg-covid-19/pull/388/files
Data download and evaluation
Dataset download: https://kg-hub.berkeleybop.io/kg-covid-19/current/kg-covid-19.nt.gz
Endpoint: http://10.254.146.165:8891/sparql
Query for evaluation:
prefix rdfs: <http://www.w3.org/2000/01/rdf-schema#>
prefix rdf: <http://www.w3.org/1999/02/22-rdf-syntax-ns#>
prefix bl: <https://w3id.org/biolink/vocab/>
prefix xsd: <http://www.w3.org/2001/XMLSchema#>
SELECT ?drug ?pred ?target WHERE
{
?drug ?pred ?target .
?drug bl:category ?drugcat .
?target bl:category ?targetcat; <https://www.example.org/UNKNOWN/ncbi_taxid> "9606"^^xsd:string .
OPTIONAL { ?drug rdfs:label ?druglab } .
OPTIONAL { ?target rdfs:label ?targetlab } .
FILTER ( ?drugcat = bl:Drug || ?drugcat = bl:ChemicalSubstance )
FILTER ( ?targetcat = bl:Protein || ?targetcat = bl:Gene )
FILTER ( ?pred = bl:molecularly_interacts_with || ?pred = bl:interacts_with )
}Rankings
RankingNew submission
To submit, create a compressed (gzip or zip) tab-separated file containing (subject, predicate, object, score) quadruples. An example file could be:
<https://reactome.org/content/detail/R-HSA-2172127> <http://purl.obolibrary.org/obo/RO_0000057> <http://www.ncbi.nlm.nih.gov/gene/5710> 0.9 <http://purl.obolibrary.org/obo/CHEBI_4034> <http://purl.obolibrary.org/obo/RO_0002434> <https://uswest.ensembl.org/Homo_sapiens/Transcript/Summary?t=ENST00000546491> 0.1 <http://purl.obolibrary.org/obo/CHEBI_49662> <http://purl.obolibrary.org/obo/RO_0002434> <https://uswest.ensembl.org/Homo_sapiens/Transcript/Summary?t=ENST00000344604> 0.231 <http://purl.obolibrary.org/obo/PR_P63010> <http://purl.obolibrary.org/obo/RO_0002436> <http://purl.obolibrary.org/obo/PR_P51812> 0.111 <http://purl.obolibrary.org/obo/MONDO_0009452> <http://purl.obolibrary.org/obo/RO_0002200> <http://purl.obolibrary.org/obo/HP_0001290> 0 <http://purl.obolibrary.org/obo/CHEBI_46195> <http://purl.obolibrary.org/obo/RO_0002434> <https://uswest.ensembl.org/Homo_sapiens/Transcript/Summary?t=ENST00000546299> 1.0 <http://purl.obolibrary.org/obo/PR_Q8ND25> <http://purl.obolibrary.org/obo/RO_0002512> <https://uswest.ensembl.org/Homo_sapiens/Transcript/Summary?t=ENST00000320619> 0.4291 <http://www.ncbi.nlm.nih.gov/gene/2065> <http://purl.obolibrary.org/obo/RO_0002559> <https://www.ncbi.nlm.nih.gov/snp/rs115311663> 0.1 <http://purl.obolibrary.org/obo/PR_Q13480-2> <http://purl.obolibrary.org/obo/RO_0002436> <http://purl.obolibrary.org/obo/PR_P16333-1> 0.5 <http://purl.obolibrary.org/obo/CHEBI_78802> <http://purl.obolibrary.org/obo/RO_0002436> <http://purl.obolibrary.org/obo/GO_0006875> 0