PheKnowLator KG
Introduction
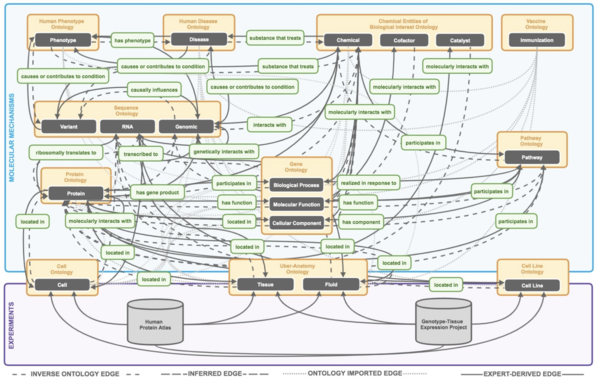
PheKnowLator (Phenotype Knowledge Translator), is a Python 3 library that constructs semantically-rich, large-scale biomedical knowledge graphs under different semantic models. For this challenge, the PheKnowLator knowledge graphs have been designed to model mechanisms of human disease and were built using 12 open biomedical ontologies, 24 linked open datasets, and results from two large-scale, experimentally-derived datasets. For additional information see the associated GitHub website: https://github.com/callahantiff/PheKnowLator/wiki/v2.0.0.
Relation types
We will evaluate predictions on the following 15 Relation Ontology (RO) relations, which are utilized in 34 distinct edge types (additional details can be found here: https://github.com/callahantiff/PheKnowLator/wiki/v2.0.0#edge-data):
| Relation | Edge Types |
|---|---|
| Participates in (http://purl.obolibrary.org/obo/RO_0000056) | chemical-pathway; gene-pathway; protein-gobp; protein-pathway |
| Has function (http://purl.obolibrary.org/obo/RO_0000085) | pathway-gomf; protein-gomf |
| Located in (http://purl.obolibrary.org/obo/RO_0001025) | protein-anatomy; protein-cell; protein-gocc; rna-anatomy; rna-cell |
| Has component (http://purl.obolibrary.org/obo/RO_0002180) | pathway-gocc |
| Has phenotype (http://purl.obolibrary.org/obo/RO_0002200) | disease-phenotype |
| Has gene product (http://purl.obolibrary.org/obo/RO_0002205) | gene-protein |
| Interacts with (http://purl.obolibrary.org/obo/RO_0002434) | chemical-gene; chemical-protein; chemical-rna |
| Genetically interacts with (http://purl.obolibrary.org/obo/RO_0002435) | gene-gene |
| Molecularly interacts with (http://purl.obolibrary.org/obo/RO_0002436) | chemical-gobp; chemical-gocc; chemical-gomf; protein-catalyst; protein-cofactor; protein-protein |
| Transcribed to (http://purl.obolibrary.org/obo/RO_0002511) | gene-rna |
| Ribosomally translates to (http://purl.obolibrary.org/obo/RO_0002513) | rna-protein |
| Causally influences (http://purl.obolibrary.org/obo/RO_0002566) | variant-gene |
| Is substance that treats (http://purl.obolibrary.org/obo/RO_0002606) | chemical-disease; chemical-phenotype |
| Causes or contributes to condition (http://purl.obolibrary.org/obo/RO_0003302) | gene-disease; gene-phenotype; variant-disease; variant-phenotype |
| Realized in response to (http://purl.obolibrary.org/obo/RO_0009501) | gobp-pathway |
Data sources and contact
In addition to the SPARQL Endpoint and RDF download link, we also provide access to the data in other formats (n-triples, networkx MultiDigraph, txt files) via a publicly accessible Google Cloud Storage Bucket. See the README in the bucket for additional documentation.
- Github: https://github.com/callahantiff/PheKnowLator
- Primary contact: Tiffany Callahan
Data download and evaluation
Dataset download: https://storage.googleapis.com/pheknowlator/ISMB_Challenge_Data/PheKnowLator_v2.0.0_full_instance_inverseRelations_noOWL_OWLNETS.nt
Endpoint: http://sparql.pheknowlator.com/sparql
Query for evaluation:
PREFIX obo: <http://purl.obolibrary.org/obo/>
PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>
SELECT ?s ?p ?o
WHERE {
VALUES ?p {
obo:RO_0000056
obo:RO_0000085
obo:RO_0001025
obo:RO_0002180
obo:RO_0002200
obo:RO_0002205
obo:RO_0002434
obo:RO_0002435
obo:RO_0002436
obo:RO_0002511
obo:RO_0002513
obo:RO_0002566
obo:RO_0002606
obo:RO_0003302
obo:RO_0009501
}
?s ?p ?o
}Rankings
RankingNew submission
To submit, create a compressed (gzip or zip) tab-separated file containing (subject, predicate, object, score) quadruples. An example file could be:
<https://reactome.org/content/detail/R-HSA-2172127> <http://purl.obolibrary.org/obo/RO_0000057> <http://www.ncbi.nlm.nih.gov/gene/5710> 0.9 <http://purl.obolibrary.org/obo/CHEBI_4034> <http://purl.obolibrary.org/obo/RO_0002434> <https://uswest.ensembl.org/Homo_sapiens/Transcript/Summary?t=ENST00000546491> 0.1 <http://purl.obolibrary.org/obo/CHEBI_49662> <http://purl.obolibrary.org/obo/RO_0002434> <https://uswest.ensembl.org/Homo_sapiens/Transcript/Summary?t=ENST00000344604> 0.231 <http://purl.obolibrary.org/obo/PR_P63010> <http://purl.obolibrary.org/obo/RO_0002436> <http://purl.obolibrary.org/obo/PR_P51812> 0.111 <http://purl.obolibrary.org/obo/MONDO_0009452> <http://purl.obolibrary.org/obo/RO_0002200> <http://purl.obolibrary.org/obo/HP_0001290> 0 <http://purl.obolibrary.org/obo/CHEBI_46195> <http://purl.obolibrary.org/obo/RO_0002434> <https://uswest.ensembl.org/Homo_sapiens/Transcript/Summary?t=ENST00000546299> 1.0 <http://purl.obolibrary.org/obo/PR_Q8ND25> <http://purl.obolibrary.org/obo/RO_0002512> <https://uswest.ensembl.org/Homo_sapiens/Transcript/Summary?t=ENST00000320619> 0.4291 <http://www.ncbi.nlm.nih.gov/gene/2065> <http://purl.obolibrary.org/obo/RO_0002559> <https://www.ncbi.nlm.nih.gov/snp/rs115311663> 0.1 <http://purl.obolibrary.org/obo/PR_Q13480-2> <http://purl.obolibrary.org/obo/RO_0002436> <http://purl.obolibrary.org/obo/PR_P16333-1> 0.5 <http://purl.obolibrary.org/obo/CHEBI_78802> <http://purl.obolibrary.org/obo/RO_0002436> <http://purl.obolibrary.org/obo/GO_0006875> 0